42 mengubah file dengan 512 tambahan dan 104 penghapusan
TEMPAT SAMPAH
save/MNIST_CNN_IID_acc.png
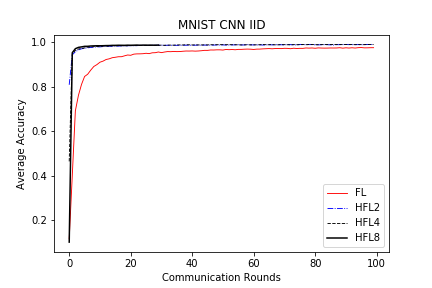
TEMPAT SAMPAH
save/MNIST_CNN_IID_loss.png
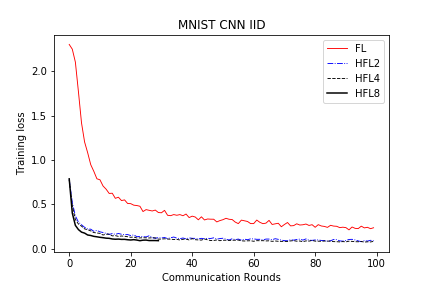
TEMPAT SAMPAH
save/MNIST_MLP_IID_acc.png
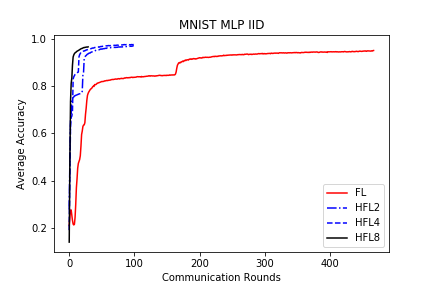
TEMPAT SAMPAH
save/MNIST_MLP_IID_loss.png
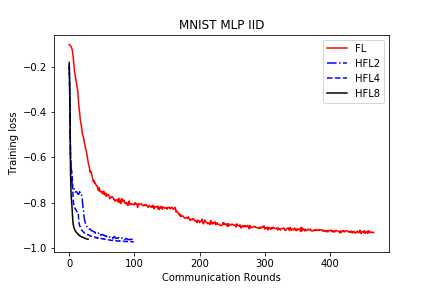
TEMPAT SAMPAH
save/MNIST_MLP_NONIID_acc.png
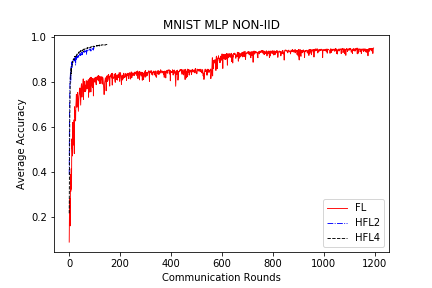
TEMPAT SAMPAH
save/MNIST_MLP_NONIID_loss.png
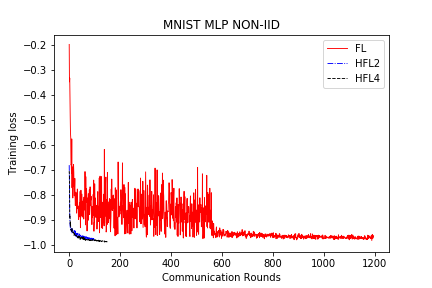
+ 0
- 0
save/objects/HFL4_mnist_mlp_100_lr[0.05]_C[0.1]_iid[0]_E[1]_B[10].pkl → save/objects/Bad/HFL4_mnist_mlp_100_lr[0.05]_C[0.1]_iid[0]_E[1]_B[10].pkl
+ 0
- 0
save/objects/HFL4_mnist_mlp_150_lr[0.05]_C[0.1]_iid[0]_E[1]_B[10].pkl → save/objects/Bad/HFL4_mnist_mlp_150_lr[0.05]_C[0.1]_iid[0]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/HFL2_mnist_mlp_100_lr[0.01]_C[0.1]_iid[1]_E[1]_B[10].pkl
+ 0
- 0
save/objects/FL_mnist_mlp_141_lr[0.1]_C[0.1]_iid[1]_E[1]_B[10].pkl → save/objects/Old/FL_mnist_mlp_141_lr[0.1]_C[0.1]_iid[1]_E[1]_B[10].pkl
+ 0
- 0
save/objects/FL_mnist_mlp_302_lr[0.1]_C[0.1]_iid[0]_E[1]_B[10].pkl → save/objects/Old/FL_mnist_mlp_302_lr[0.1]_C[0.1]_iid[0]_E[1]_B[10].pkl
+ 0
- 0
save/objects/[1]FL_mnist_mlp_200_lr[0.05]_C[0.1]_iid[1]_E[1]_B[10].pkl → save/objects/Old/[1]FL_mnist_mlp_200_lr[0.05]_C[0.1]_iid[1]_E[1]_B[10].pkl
+ 0
- 0
save/objects/[2]FL_mnist_mlp_302_lr[0.05]_C[0.1]_iid[0]_E[1]_B[10].pkl → save/objects/Old/[2]FL_mnist_mlp_302_lr[0.05]_C[0.1]_iid[0]_E[1]_B[10].pkl
+ 0
- 0
save/objects/[3]HFL2_mnist_mlp_101_lr[0.05]_C[0.1]_iid[1]_E[1]_B[10].pkl → save/objects/Old/[3]HFL2_mnist_mlp_101_lr[0.05]_C[0.1]_iid[1]_E[1]_B[10].pkl
+ 0
- 0
save/objects/[4]HFL2_mnist_mlp_101_lr[0.05]_C[0.1]_iid[0]_E[1]_B[10].pkl → save/objects/Old/[4]HFL2_mnist_mlp_101_lr[0.05]_C[0.1]_iid[0]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/Old/clustersize50HFL4_mnist_mlp_100_lr[0.01]_C[0.1]_iid[1]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/[10]FL_mnist_cnn_261_lr[0.01]_C[0.1]_iid[0]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/[11]HFL2_mnist_cnn_100_lr[0.01]_C[0.1]_iid[1]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/[12]HFL2_mnist_cnn_100_lr[0.01]_C[0.1]_iid[0]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/[13]HFL4_mnist_cnn_100_lr[0.01]_C[0.1]_iid[1]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/[15]HFL8_mnist_cnn_30_lr[0.01]_C[0.1]_iid[1]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/[16]HFL8_mnist_cnn_30_lr[0.01]_C[0.1]_iid[0]_E[1]_B[10].pkl
+ 0
- 0
save/objects/FL_mnist_mlp_468_C[0.1]_iid[1]_E[1]_B[10].pkl → save/objects/[1]FL_mnist_mlp_468_C[0.1]_iid[1]_E[1]_B[10].pkl
+ 0
- 0
save/objects/FL_mnist_mlp_1196_lr[0.01]_C[0.1]_iid[0]_E[1]_B[10].pkl → save/objects/[2]FL_mnist_mlp_1196_lr[0.01]_C[0.1]_iid[0]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/[3]HFL2_mnist_mlp_100_lr[0.01]_C[0.1]_iid[1]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/[4]HFL2_mnist_mlp_100_lr[0.01]_C[0.1]_iid[0]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/[5]HFL4_mnist_mlp_100_lr[0.01]_C[0.1]_iid[1]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/[6]HFL4_mnist_mlp_150_lr[0.01]_C[0.1]_iid[0]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/[7]HFL4_mnist_mlp_30_lr[0.01]_C[0.1]_iid[1]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/[9]FL_mnist_cnn_100_lr[0.01]_C[0.1]_iid[1]_E[1]_B[10].pkl
TEMPAT SAMPAH
save/objects/clustersize50HFL4_mnist_mlp_100_lr[0.01]_C[0.1]_iid[0]_E[1]_B[10].pkl
File diff ditekan karena terlalu besar
+ 143
- 0
src/.ipynb_checkpoints/Eval-checkpoint.ipynb
File diff ditekan karena terlalu besar
+ 16
- 12
src/.ipynb_checkpoints/federated-hierarchical_v1_twoclusters-changeEval-checkpoint.ipynb
+ 209
- 0
src/Eval.ipynb
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
TEMPAT SAMPAH
src/__pycache__/models.cpython-37.pyc
+ 25
- 8
src/baseline_main.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 6
- 6
src/federated-hierarchical2_main.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 19
- 19
src/federated-hierarchical4_main.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 33
- 35
src/federated-hierarchical8_main.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
File diff ditekan karena terlalu besar
+ 33
- 17
src/federated-hierarchical_v1_twoclusters-changeEval.ipynb
+ 2
- 1
src/federated_main.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
+ 26
- 6
src/models.py
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||
|
||